Back to top
PCR Probes

Increase the sensitivity and specificity of your real-time or digital PCR with probes from Microsynth. High-quality qPCR and dPCR probes with a broad range of dyes and quenchers are delivered at exceptional speed.
Features and Benefits
-
Thorough purification processes (HPLC or even PAGE)
-
Stringent quality control (online trityl monitoring and MALDI-TOF MS)
- Within 3 to 5 working days
User-friendly Online Ordering System
-
Easy-to-use online portal with a series of helpful tools (e.g. order tracking & history, convenient search and re-order option)
- Possibility of adjusting binding affinity via MGB, LNA and other Tm enhancers or even combining different Tm enhancers to specifically tailor your assay
- Wide variety of fluorophores - quencher combinations
- Professional design service
- EN ISO 13485:2016 certified production process
- qPCR & digital PCR assay development, validation, manufacturing, and testing can be outsourced to Microsynth
- Our highly experienced and well-trained scientists are happy to support you.
Dual-Labeled Probes
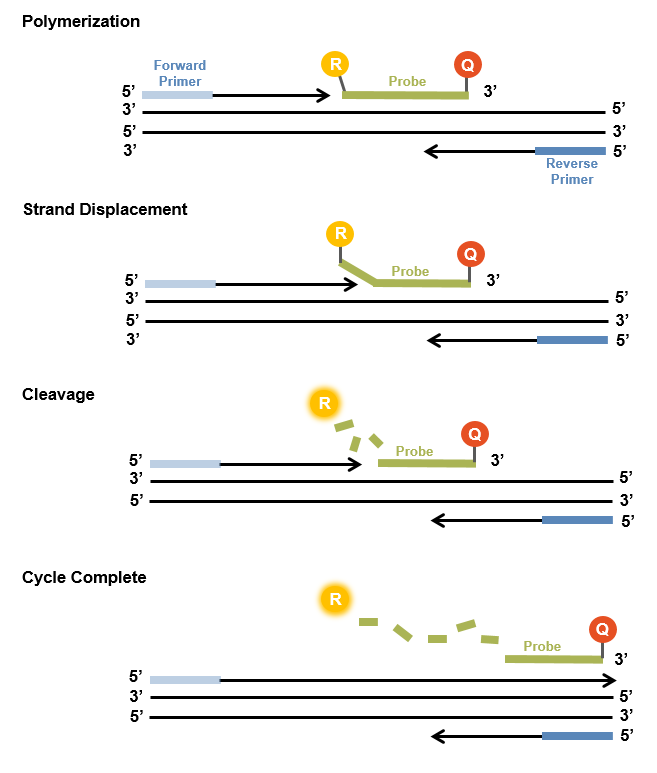
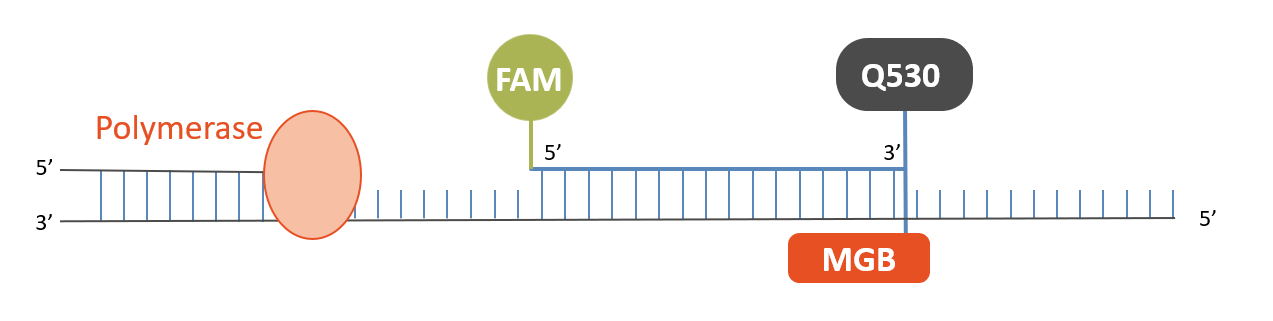
The so-called TaqMan probes are hydrolysis probes that are designed to increase the specificity of quantitative PCR.
In the hybridized state the proximity of the fluorophore and the quencher is sufficient to quench the fluorescence. During the PCR the 5’-3’ exonuclease activity of the Taq polymerase is used to cleave the dual-labeled probe. In that way, the fluorophore and the quencher are separated and a fluorescence signal permits quantitative measurement.
The TaqMan probe significantly increases the specificity of detection.
We'd be delighted to assist you with the design of the probes and the associated primers.
| 5‘ Label | 3‘ Label | 0.04 µmol scale1,2,3 | 0.2 µmol scale1,2,3 | 1.0 µmol scale1,2,3 |
|
FAM |
BHQ-1 |
1 OD |
2 OD 10 nmol |
12 OD 60 nmol |
|
AlexaFluor Dyes |
BHQ-1 or BHQ-2 |
0.75 OD 3.75 nmol |
1.5 OD 7.5 nmol |
8 OD 40 nmol |
2 Yields indicated in OD260 apply to a probe consisting of 20 DNA bases; Calculation: 1 OD = 5 nmol (please note that this calculation is based on sequences with virtually homogeneous distribution of the 4 DNA bases; it may vary for sequences with high GC contents >70% etc.)
3 Guaranteed yields for PAGE purified Oligos are lower
Double-Quenched Probes
The usual qPCR probes for 5'-nuclease assays have a fluorophore at the 5' end and a suitable quencher at the 3' end. On the one hand, this clears the fluorescence of the dye while on the other hand, it prevents the prolongation of the probe during the qPCR reaction. For longer probes and thus a greater distance between fluorophore and quencher, the background often increases to an undesirable degree.
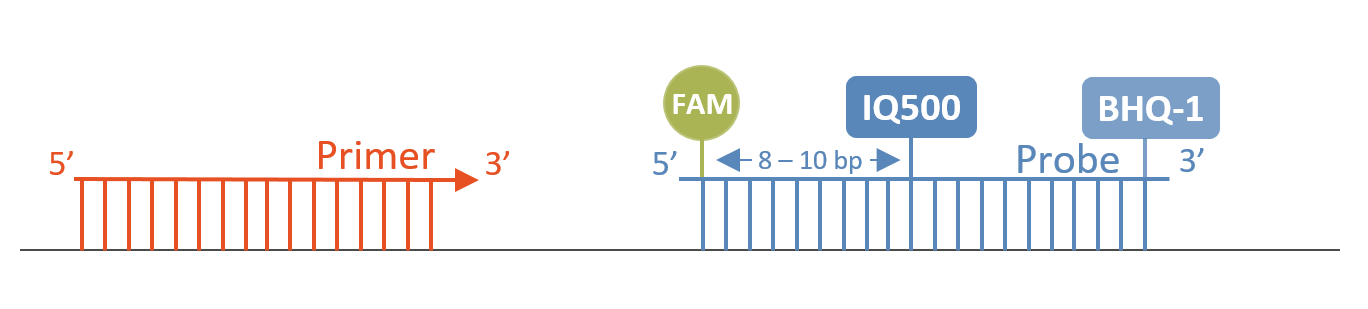
Double-quenched probes reduce the background and increase the sensitivity. By incorporating an additional quencher into the sequence near the 5'-fluorophore, a significant improvement can be realized in the results.
The new quencher IQ-500 with an absorption maximum of approx. 500 nm and a quenching range of approx. 450 nm to 550 nm is designed as a 3' and internal modification.
Double-quenched probes contain a 5'-FAM fluorophore, a 3'-BHQ-1 quencher and an internal IQ-500 quencher at a distance of 8-10 bases from the fluorophore.
We's be delighted to assist you with the design of the probes and the associated primers.
| 5‘ Label | 3‘ Label | 0.04 µmol scale1,2 | 0.2 µmol scale1,2 | 1.0 µmol scale1,2 |
| FAM | IQ-500 with BHQ-1 or TAMRA |
1 OD |
2 OD 10 nmol |
8 OD 40 nmol |
1 The synthesis scale represents the initial amount of 3' bases (starting material).
2 Yields indicated in OD260 apply to a probe consisting of 20 DNA bases; Calculation: 1 OD = 5 nmol (please note that this calculation is based on sequences with virtually homogenous distribution of the 4 DNA bases; it may vary for sequences with high GC contents >70% etc.)
Potential Applications
- Demanding qPCR applications with a need for greater flexibility in sequence design, e.g. for targets that are AT-rich and require longer probes (≥25 bases)
- ddPCR analysis where the double-quenched probe facilitates discrimination of positive vs negative droplets (improved cluster)
LNA Probes
Locked Nucleic Acid (LNA) has proven to be a powerful tool in many qPCR and digital PCR applications. When incorporated into DNA oligonucleotides, the resulting LNA containing oligonucleotides offer the following main benefits compared to native-state DNA bases only:
- Increased thermal stability and hybridization specificity
- More accurate gene quantification and allelic discrimination
- Easier and more flexible designs for problematic target sequences
Each LNA base addition in an oligo increases the Tm by approximately 2-4 °C and locked nucleic acid probes can be designed to have a ΔTm of >15°C. Usually, no more than 5 LNA bases are incorporated into a probe sequence. When designing and optimizing your LNA probes, please ensure you follow the guidelines provided here.
Microsynth offers the opportunity for you to order your LNA probe with our full range of dyes and quenchers and this also applies to our dual labeled probes.
| 0.04 µmol scale1,2 | 0.2 µmol scale1,2 | 1.0 µmol scale1,2 | ||
| LNA probes |
0.5 OD |
1 OD 5 nmol |
6 OD 30 nmol |
|
MGB Probes
- Higher target binding selectivity
- Reduced background fluorescence
- Enhanced quality of the probe due to shorter length
| 5‘ Label | 3‘ Label | 0.04 µmol scale1,2,3 | 0.2 µmol scale1,2,3 | 1.0 µmol scale1,2,3 |
| FAM JOE Yakima Yellow HEX |
MGB-Q5304 |
1 OD
5 nmol
|
2 OD 10 nmol |
10 OD 50 nmol |
2 Yields indicated in OD260 apply to a probe consisting of 20 DNA bases; Calculation: 1 OD = 5 nmol (please note that this calculation is based on sequences with virtually homogenous distribution of the 4 DNA bases; it may vary for sequences with high GC contents >70% etc.)
4 In addition, Microsynth offers the cost-effective 3’ MGB-Q500 quencher moiety. This quencher is only available with FAM as reporter dye and may represent a good entry. For higher demands (top quenching, multiplexing) 3’ MGB-Q530 is the quencher of choice.
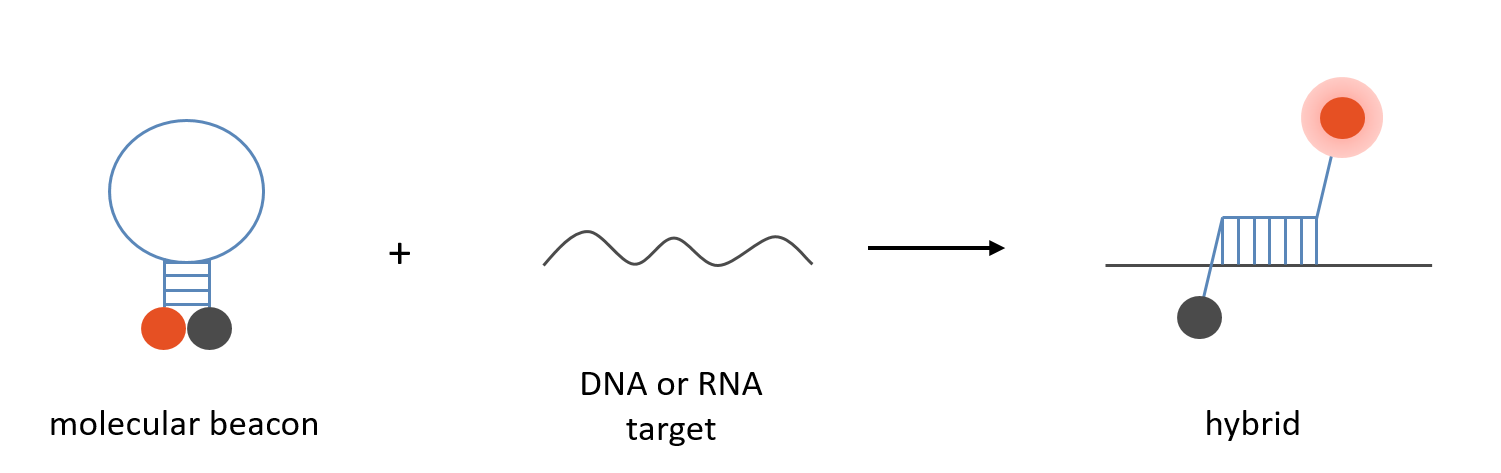
Molecular Beacons
How to Order
- Enter our webshop
- Click on DNA in the "DNA/RNA Synthesis" domain
- Select either Normal Entry in order to type or copy/paste the desired sequence information etc. or alternatively select Upload Entry by using our convenient Excel template (can be downloaded during ordering)
- Single-quenched probes (TaqMan, Molecular Beacon, MGB and LNA probes): Select your desired 5', 3' modification as well as HPLC purification1, and your oligo is recognized and processed as a single-quenched probe.
- In case of LNA probes include the ordering symbols “5, 6, 7, and 8” into your sequence and choose “LNA-A, LNA-C, LNA-G, and LNA-T” under “Inner Modification (5=…)” etc.
- Double-quenched probes: Include ordering symbol "5" into your sequence (e.g. catattgaa5actgggttaacggaatt) and choose "Internal Quencher 500" under Inner Modification ("5=…"). Select your desired 5', 3' modification as well as HPLC purification, and your oligo is recognized and processed as a double-quenched probe.
- Follow the further instructions
1 Microsynth delivers qPCR probes of the highest quality. The majority of Microsynth qPCR probes are purified by HPLC whereas a few require PAGE purification in order to meet the stringent QC specifications. If your dye/quencher combination is not compatible with HPLC just use PAGE.